2026 Virtual Genome Writing Workshop
The Dark Matter Project is holding a free online training series on the application of synthetic genomics (genome writing) in mammalian cells.
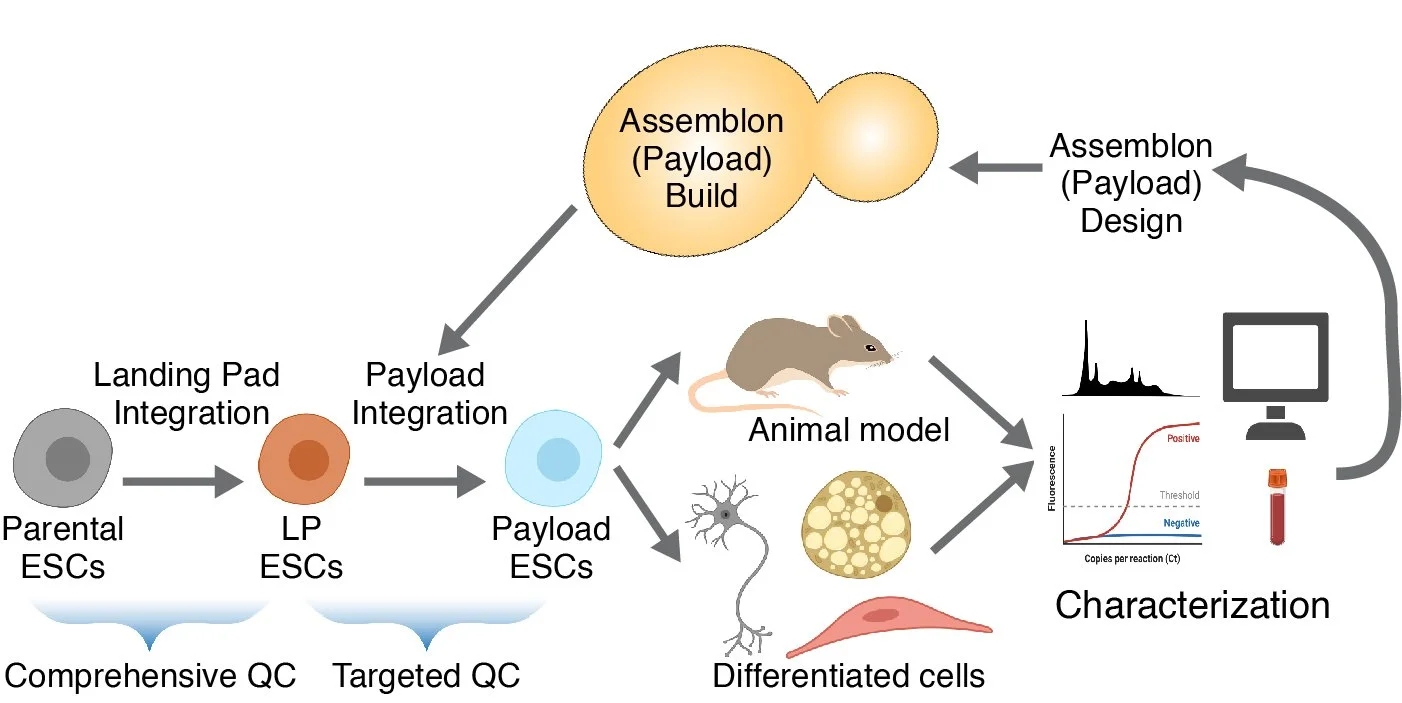
The series will include one introductory session (open to all) in which we’ll highlight the applications and (our) technologies for building Big DNAs, delivering them to mammalian genomes, comprehensive sequence verification and readout , as their bioethical considerations of genome writing.
Two additional sessions will be offered for a limited number of participants in which we will live-demo the design process for mammalian genome writing, including how to select the boundaries of an engineered locus and how to design gRNAs expression vectors, landing pads, Big DNA constructs, genotyping strategies and more. We will highlight the use of the UCSC genome browser, BLAST and SnapGene as efficient tools for this design process.
Registration for Sessions 2-4 is no closed.
You can still register to Session 1
Format and dates (agenda will be shared with participants):
Session 1 (open to all) — Introductory Webinar — March 16, 2026, 10:00 am - 12:30 pm (ET):
Applications of mammalian genome writing (Dr. Jef Boeke)
Introduction to mammalian genome writing technologies — Big DNA assembly, delivery, sequence-verification and readout (Dr. Jef Boeke, Dr. Ran Brosh, Dr. Raquel Ordoñez and Jack Atwater).
Genome writing bioethics considerations (Dr. Carolyn Riley Chapman)
Sessions 2 and 3 (limited participation) — Virtual Mammalian Genome Writing Design — March 23 and March 30, 2026, 10:00 am - 12:30 pm (ET).
Led by Dr. Ran Brosh:
Live online demonstration: using the UCSC genome browser, SnapGene and BLAST for efficient and comprehensive design of genome writing strategies
Choosing locus boundaries
Designing gRNAs and expression vectors
Designing landing pads and Big DNA payloads
Designing PCR genotyping strategies
Considerations for genome writing verification
Session 4 (for graduates of sessions 1-3) — Design-Your-Own-Locus — April 13, 2026, 10:00 am - 12:30 pm (ET):
Participants would be given an opportunity to design their own genome writing strategy of their favorite gene/locus, guided by one of the Dark Matter Project’s genome writing experts.
Prerequisites:
Participation in the live-demo Sessions 2-4 is limited to to trainees, faculty and R&D scientists
Participants must be familiar with DNA cloning, CRISPR and mammalian cell and molecular biology
Participants must be familiar with the DNA cloning design software SnapGene. Participation in Session 4 requires a fully activated SnapGene license (which we cannot provide, but a 30-day free trial is offered by SnapGene for first-time users.
Participants must be familiar with the UCSC Genome Browser and with BLAT/BLAST
Suggested reading:
Pinglay et al., Mammalian genome writing: Unlocking new length scales for genome engineering. Cell. 2026.
Brosh et al., A versatile platform for locus-scale genome rewriting and verification. Proceedings of the National Academy of Sciences. 2021.
Zhang et al., Mouse genome rewriting and tailoring of three important disease loci. Nature. 2023.
Brosh et al., Synthetic regulatory genomics uncovers enhancer context dependence at the Sox2 locus. Mol Cell. 2023.
Applications
Three of the many applications of Big DNA
Our genome writing pipeline
From stem cells to data and back again