Assemble libraries of >100 kilobases synthetic constructs (Assemblons) harboring systematic variation in regulatory elements for multiple gene loci.
For each locus of interest, we build a library of full-length alleles (including regulatory regions) ranging upwards of 100 kb. As these are constructed by homologous recombination-based assembly in yeast, we refer to these as “assemblons”.
There are three main types of mammalian “assemblon” projects (see Projects):
Synthetic haplotypes – are a rapid way to dissect a risk haplotype associated with disease susceptibility.
Synthetic hypervariation – a way to probe the network of regulatory sites controlling a gene of interest by manipulation of potential enhancer and promoter elements.
Humanized mice – large fragments of human DNA, usually containing a gene with its entire regulatory regions, which is then “transplanted” into the mouse genome in order to generate human disease models.
We use several types of source DNA for our assemblons:
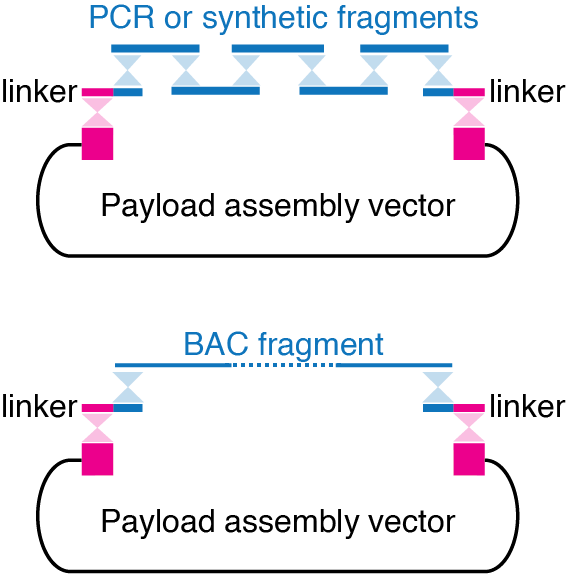
Synthetic DNA (examples: Backwards DNA)
Fragments of human or mouse BACs (bacterial artificial chromosomes) that we digest with CRISPR/Cas9 (examples: Sox2, H19/Igf2)
Assembly of all the fragments is usually mediated by short synthetic “linker” DNA fragments or by terminal homologies “encoded” directly in the fragments.
After building these “base” assemblons we often generate a series of variant assemblons using a variety of methods, but mostly by employing CRISPR technology to introduce changes (deletions, inversions, relocations, mutation etc.) to the base assemblon in yeast. Read more about how we developed a complete toolset for this purpose, which we call CREEPY (CRISPR Engineering of EPisomes in Yeast) in Zhao et al., bioRxiv. 2022.
Our Synthesis and Assembly Technology:
We have mastered the technology of assembling very large DNAs using the power of homologous recombination in yeast. A critical resource facilitating these activities is the GenomeFoundry@ISG, which has a robotic workcell designed specifically to automate one of the most central and high cost activities in assembling long DNAs: verification of complete and correct assemblies. Our automated screening protocol (Mitchell et al., JoVE 2015) was adapted to identify yeast colonies bearing Assemblons of the right structure in very high throughput (Mitchell et al., bioRxiv 2018).
We have also developed a LIMS-based online system called MenDEL for the purpose of Assemblon design as well as managing the substantial data and metadata transactions associated with constructing each Assemblon.
Our advanced synthesis and assembly technologies have already been successfully translated in the field of synthetic biology. We reported the first complete “designer” yeast chromosome arm in 2011 (Dymond et al., Nature 2011) and total synthesis of the first synthetic eukaryotic chromosome in 2014 (Annaluru et al., Science 2014). We lead a worldwide Synthetic Yeast 2.0 (Sc2.0) consortium aimed at producing a fully synthetic (and modified) yeast genome. In 2017 we reported the complete synthesis of several yeast chromosomes (Mitchell et al., Science 2017; Shen et al., Science 2017; Wu et al., Science 2017; Xie et al., Science 2017; Zhang et al., Science 2017, bioRxiv 2022), including a megabase chromosome, and the complete design of the entire genome (Richardson et al., Science 2017), as well as the first reports on the 3D structure of synthetic chromosomes (Mercy et al., Science 2017). More recently we consolidated many of these synthetic chromosomes to a single yeast stain (Zhao et al., bioRxiv 2022).